BUFFALO, NY- May 19, 2022 – A new research paper was published in Volume 13 of Oncotarget, entitled, “CancerOmicsNet: a multi-omics network-based approach to anti-cancer drug profiling.”
Researchers from Louisiana State University developed CancerOmicsNet—a graph neural network model to predict the growth rate of a cancer cell line after drug treatment.
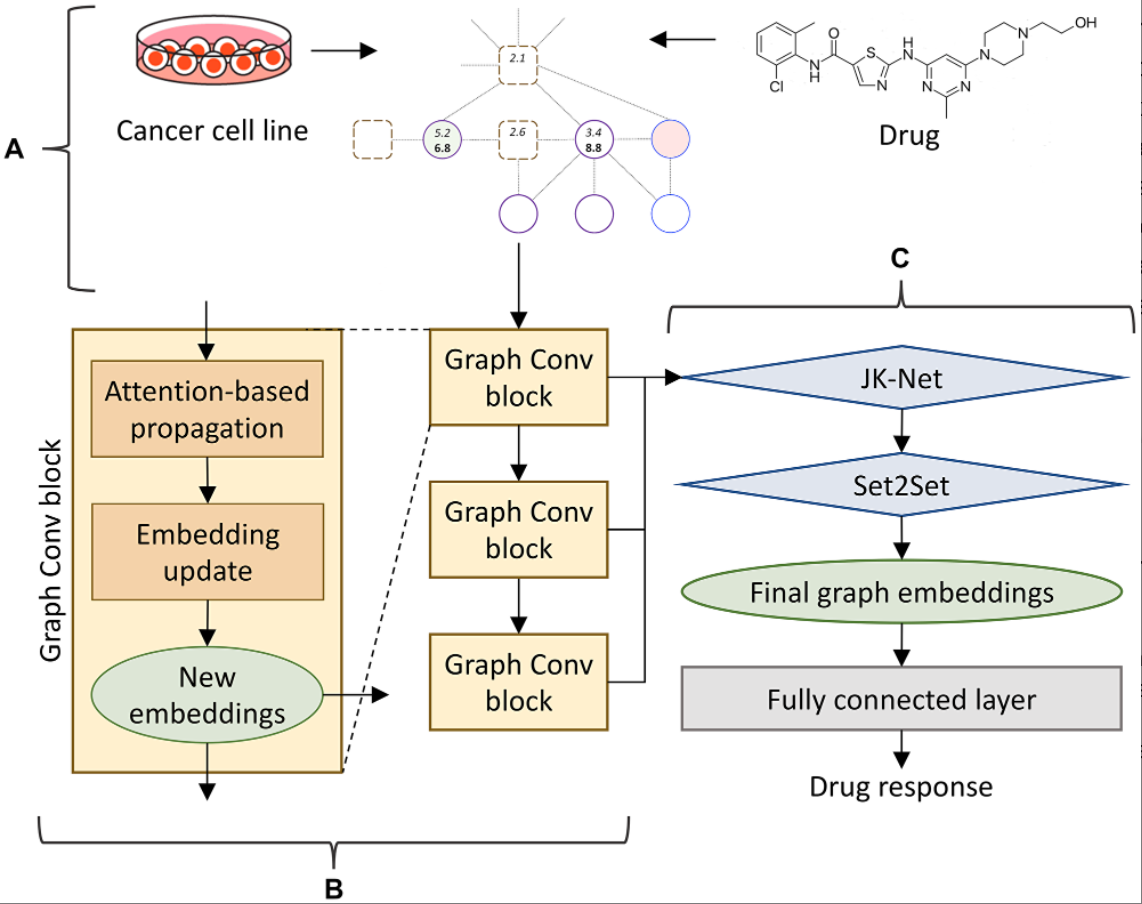
“CancerOmicsNet is more advanced than many deep learning techniques operating in the Euclidean space [47], because it extracts knowledge directly from biological networks providing a more adequate representation of complex diseases such as cancer.”
CancerOmicsNet integrates multiple heterogeneous data, such as biological networks, genomics, inhibitor profiling, and gene-disease associations, into a unified graph structure. This novel method utilizes compact, cancer-specific networks constructed from protein-protein interactions, differential gene expression, disease-gene association, and drug inhibition data.
“In order to evaluate the performance of CancerOmicsNet, we conducted a cross-validation at the tissue level by removing from model training all cell lines originating from a particular tissue and then analyzing the accuracy for these cell lines.”
In this study, the researchers carefully evaluated the generalizability of CancerOmicsNet in a series of cross-validation benchmarks against different tumor tissues. Encouragingly, the cross-validated accuracy of CancerOmicsNet at the tissue level was significantly higher than those measured for other approaches on the same data.
“The performance of CancerOmicsNet, properly cross-validated at the tissue level, is 0.83 in terms of the area under the receiver operating characteristics, which is notably higher than those measured for other approaches.”
The researchers specify that the applicability of CancerOmicsNet is presently limited to kinase inhibitors, while alternative methods are applicable to other classes of therapeutics as well.
“Overall, CancerOmicsNet offers a high performance and the desired generalizability in the prediction of the effect of kinase-targeted therapies on the cancer cell growth.”
DOI: https://doi.org/10.18632/oncotarget.28234
Correspondence to: Michal Brylinski – Email: michal@brylinski.org
Keywords: cancer growth rate, kinase inhibitors, differential gene expression, gene-disease association, cancer-specific networks
ONCOTARGET VIDEOS: YouTube | LabTube | Oncotarget.com
About Oncotarget
Oncotarget (a primarily oncology-focused, peer-reviewed, open access journal) aims to maximize research impact through insightful peer-review; eliminate borders between specialties by linking different fields of oncology, cancer research and biomedical sciences; and foster application of basic and clinical science.
To learn more about Oncotarget, visit Oncotarget.com and connect with us on social media:
- Twitter – https://twitter.com/Oncotarget
- Facebook – https://www.facebook.com/Oncotarget
- YouTube – www.youtube.com/c/OncotargetYouTube
- Instagram – https://www.instagram.com/oncotargetjrnl/
- LinkedIn – https://www.linkedin.com/company/oncotarget/
- Pinterest – https://www.pinterest.com/oncotarget/
- LabTube – https://www.labtube.tv/channel/MTY5OA
- SoundCloud – https://soundcloud.com/oncotarget
For media inquiries, please contact: media@impactjournals.com.