Mariana Perepitchka, Yekaterina Galat, and Dr. Vasiliy Galat discuss their 2020 research paper published by Oncotarget, entitled, “Down syndrome iPSC model: endothelial perspective on tumor development.”
Behind the Study is a series of transcribed videos of researchers elaborating on their oncology-focused research published by Oncotarget. Visit the Oncotarget YouTube channel for more insights from outstanding authors.
—
Hello, my name is Mariana Perepitchka and I am a research associate at the Galat Laboratory which is located at the Simpson Querrey Biomedical Research Center and affiliated with Ann and Robert H. Lurie Children’s Hospital and Northwestern University Feinberg School of Medicine. On behalf of all the authors, I would like to say thank you for the opportunity to present this research.
Hello, my name is Yekaterina Galat. I am also a research associate in Galat Laboratory. Together with Mariana, we are the lead coauthors on our paper titled Down syndrome induced pluripotent stem cell model in epithelial insights into tumor development (published on Oncotarget.com).
My name is Vasiliy Galat, and I am head of the lab. My lab has a long-standing interest in disease modeling. In fact, it was the first to establish embryonic stem cells from human embryos, with genetic disease. These days, we are using reprogramming technology to develop patient specific induced pluripotent stem cells, or IPCs. These stem cells can be differentiated into different type of cells and used for research and testing. This concept is known as a disease in the dish.
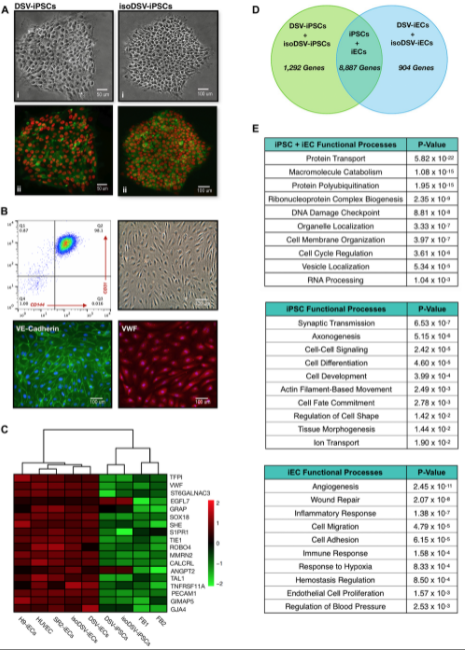
Our laboratory has several ongoing project in role of hematopoietic differentiation. For instance, we study natural killer cells, but use from IPCs for immune therapy applications. In this particular paper, we are presenting our investigation of iPSCs, direct endothelial cells. Endothelial cells play a pivotal role in blood formation. And we choose them to study trisomic chromosome 21 implication on leukemia development in down syndrome patients.
Yekaterina Galat
Endothelial cells are not easily attainable from patients. This is why our endothelial iPSC model proves to be so useful. While deriving endothelial cells from down syndrome patients, we noticed that down syndrome proliferative response to vascular endothelial growth factor, differed from control IPCs.
This finding, coupled with clinical data showing that down syndrome patients have high incidents of leukemia and decreased solid tumor prevalence, motivated us to further analyze this down syndrome endothelial progenitors. For this study, we performed endothelial derivation, bioinformatic analysis and functional assays.
Mariana Perepitchka
Regarding the bioinformatic component, we utilized various cancer databases and carried out extensive pathway analysis. The functional essays targeted proliferation, migration and inflammatory response, which are processes that tumors employ to create a favorable niche for growth in order to transition into a more invasive phenotype.
Our analysis revealed several notable aspects. For example, we observed an increased expression in leukemia associated oncogenes. So for example, GATA2 and NOTCH1. And decreased expression of genes involved in regulating inflammation, such as IL-6, IL-8, CCL2.
And we believe that this genetic profile is contributing toward a leukemia conducive microenvironment. And importantly, most of our identified endothelial genes have not been previously highlighted in down syndrome research, which is predominantly centered around genes expressed on chromosome 21.
Yekaterina Galat
I would say that one of the most challenging aspects about this study was the optimization of endothelial denervation to ensure that our endothelial progenitors demonstrate the appropriate molecular and phenotypic characteristic. At the time when this study was initiated, inefficient endothelial derivation method was not available. As a result, we dedicated a significant amount of our time to establishing the robust endothelial differentiation protocol.
Mariana Perepitchka
A surprising aspect of this study was that the upregulated leukemia associated oncogenes and the differentially expressed genes linked to proliferation, migration and inflammatory response, were actually not located on chromosome 21. And this significant finding emphasizes how trisomy 21 is capable of leading genome-wide alterations that result in down syndrome endothelial dysfunction and increased leukemic susceptibility.
We actually observed similar genome-wide dysregulation in a recently published parallel study that incorporates mesodermal cells. And this work shows that processes underlying connective tissue impairment in down syndrome patients, also potentially hinder solid tumor progression. So overall, down syndrome provides a unique, non-traditional lens to study tumor development.
Vasiliy Galat
We defined a list of candidate genes of interest. The next step of our investigation would be to perform in vivo verification of these genes. We may have new gene targets for leukemia treatment, which is very exciting. Thank you for your attention.
Yekaterina Galat
Please read our paper and contact us with any questions you may have.
Mariana Perepitchka
Thank you again.
Click here to read the full research paper published by Oncotarget.
ONCOTARGET VIDEOS: YouTube | LabTube | Oncotarget.com
—
Oncotarget is a unique platform designed to house scientific studies in a journal format that is available for anyone to read—without a paywall making access more difficult. This means information that has the potential to benefit our societies from the inside out can be shared with friends, neighbors, colleagues and other researchers, far and wide.
For media inquiries, please contact media@impactjournals.com.